Creating a fully-featured database was unfeasible. Instead, we launched the free tool, AlphaFold Server, which allows scientists to input their own sequences for AlphaFold to generate molecular complexes. Since its release in May, researchers have used it to generate over a million structures.
"It’s like Google Maps for molecular complexes," says Lindsay Willmore, a research engineer at Google DeepMind. "Anyone without coding knowledge can simply copy and paste their protein, DNA, RNA sequences, or small molecule names, click a button, and wait a few minutes. The structure and confidence measures will be revealed for them to examine and evaluate their predictions."
To enable AlphaFold 3 to handle a broader range of biomolecules, the team significantly expanded the training data to include DNA, RNA, small molecules, and more. "We decided to train on everything available in this dataset that helped us so much with proteins and see how far we could go," Lindsay explains. "And it turns out, we can go quite far."
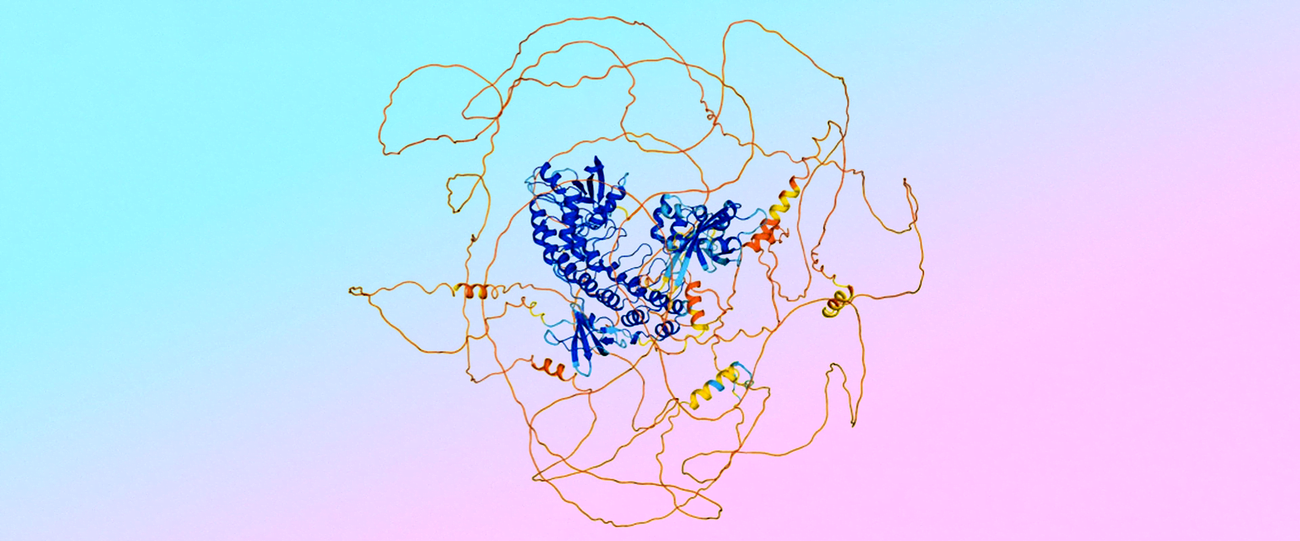
A major change in AlphaFold 3 is the architectural shift in the final model that generates the structure. While AlphaFold 2 used a complex module based on custom geometry, AlphaFold 3 employs a diffusion-based generative model, similar to our advanced image generation models like Imagen. This greatly simplified how the model handles new types of molecules.
However, this change introduced a new problem: the diffusion model would inaccurately create an "ordered" structure with a defined spiral shape for the so-called "disordered regions" of proteins, as these regions were not included in the training data.
The team turned to AlphaFold 2, which is highly effective at predicting which interactions would be disordered—those resembling a chaotic pile of spaghetti—and which would not. "We used these structures predicted by AlphaFold 2 as distillation training for AlphaFold 3, so AlphaFold 3 could learn to predict disorder," Lindsay explains.
"We have a saying: ‘Trust the fusilli, reject the spaghetti,’" adds Jonas.